Our Research
Our laboratory focuses on a fundamental biological question of how precursor cells (i.e., stem cells and progenitors) are epigenetically regulated to support healthy tissue formation, regeneration and homeostasis. By tackling this question with modern genetic, biochemical, molecular biological and genomic means, we hope to uncover new insights into understanding the molecular mechanisms and functional consequences of tumorigenesis.
My lab has developed a new system by which we have identified key epigenetic regulators that control progenitor expansion and differentiation. Our recent work has also identified non-coding RNAs as players modulating the epigenetic property of progenitors. We hope our research will result in therapeutic implications for boosting tissue regeneration and ameliorating tumor progression.
My lab has developed a new system by which we have identified key epigenetic regulators that control progenitor expansion and differentiation. Our recent work has also identified non-coding RNAs as players modulating the epigenetic property of progenitors. We hope our research will result in therapeutic implications for boosting tissue regeneration and ameliorating tumor progression.
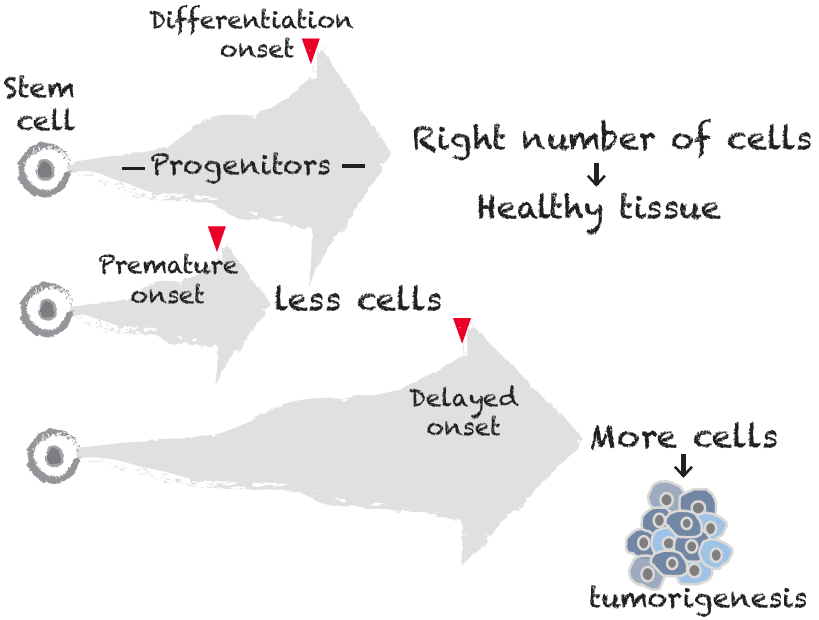
Tissue formation needs a precise timing of differentiationThe precise control over progenitor proliferation is essential for maintaining tissue homeostasis. While precocious onset of differentiation generates too few of cells and compromises tissue integrity, delayed differentiation leads to tissue overgrowth and possibly tumorigenesis.
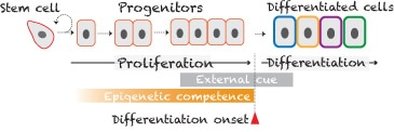
Progenitors acquire epigenetic competence to differentiate
Our recent work (Lee et.al., 2014) discovered that progenitors are likely epigenetically modified during proliferation cycles to gain responsiveness (i.e., competence) toward pre-existing external cues. Such changes of progenitors' intrinsic properties likely occur at chromatin level mediated by LSD1 (i.e., Lysine-Specific Demethylase 1) complex.
|
Our system
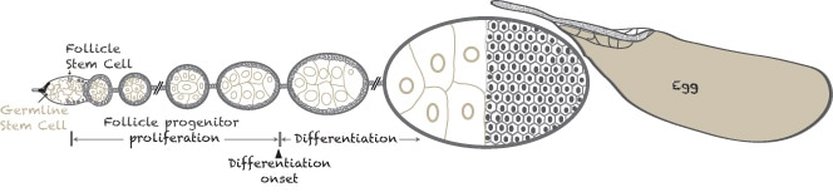
We use the fly ovarian follicle cell lineage, the longest somatic cell lineage in fly, to investigate how progenitor cells are regulated epigenetically to differentiate at the correct timing.
In the follicle cell lineage, follicle progenitors undergo 8 mitotic cycles stereotypically before differentiation.
In the follicle cell lineage, follicle progenitors undergo 8 mitotic cycles stereotypically before differentiation.
#1: Investigate the epigenetic machinery that controls proliferation & differentiation switch in progenitors
Who are the players?
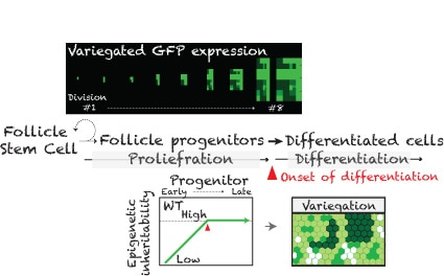
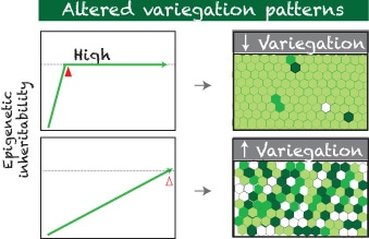
By looking for genetic mutants that show altered GFP variegation patterns, we aimed to identify genes that are required for modulating epigenetic inheritability and control progenitor differentiation.
|
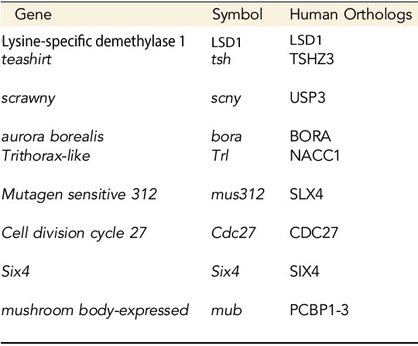
Took advantage of powerful genetic screen (haploid deficiency screening), we identified 9 genes that dominantly regulate epigenetic inheritability and progenitor proliferation. We are in the process of further investigating these candidate genes to determine their specific roles in modulating epigenetic landscapes for directing robust differentiation programs.
|
|
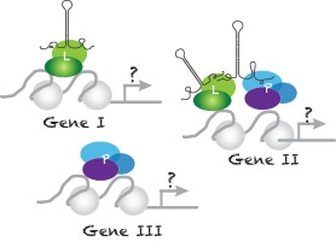
Among the genes identified, we discover that LSD1 acts together with its binding partner, CoRest, as a H3K4-specific demethylase to regulate epigenetic inheritability and functions as a master regulator repressing progenitor competence.
While early progenitors with high LSD1 expression are not responsive to Notch signaling, the reduced Lsd1 expression in late progenitors likely is required for progenitors to gain competence and respond to the pre-existing external cue, Delta, the Notch ligand. Our findings support a new model that progenitors are first prepared epigenetically to acquire competence to later respond to induction cues. Further studies investigating how lsd1 controls epigenetic inheritability in situ to modulate cellular competence will be a future research focus of our lab. |
LSD1 function as n epigenetic eraser to suppresses progenitor competence
|
#2 Epigenetic regulations that are modulated by proteins and beyond
|
Through immuno-precipitatating down non-coding RNAs (ncRNAs) that associated with LSD1, we identified a novel population of LSD1-interacting ncRNAs (LINRs) that likely modulate the function of LSD1 complex to influence epigenetic landscape in stem and progenitor cells.
We are currently investigating how these LINRs are involved in modifying the functionality, stability and targeting of LSD1 complex in vivo. |